%%html
<script src="https://bits.csb.pitt.edu/preamble.js"></script>
%%html
<div id="dtques" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#dtques';
jQuery(divid).asker({
id: divid,
question: "Is a decision tree prediction performed by asking a series of yes/no questions?",
answers: ["Yes","No","Maybe"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Example (revisited)¶
import pandas as pd
import numpy as np
import sklearn
data = pd.read_csv('compounds.fp',header=None,delim_whitespace=True)
labels = data.iloc[:,1].to_numpy()
features = data.iloc[:,2:].to_numpy()
Variance vs Bias¶
 Random forests is an example of an averaging ensemble method, where the goal is to reduce variance.
Random forests is an example of an averaging ensemble method, where the goal is to reduce variance.
Boosting ensemble methods try to reduce bias (fit the data better).
Random Forest Model Optimization¶
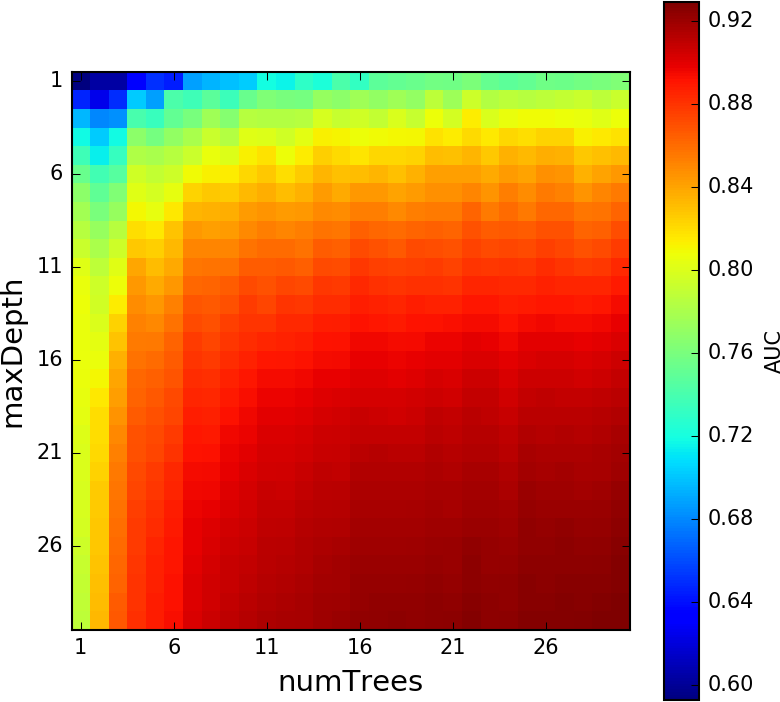
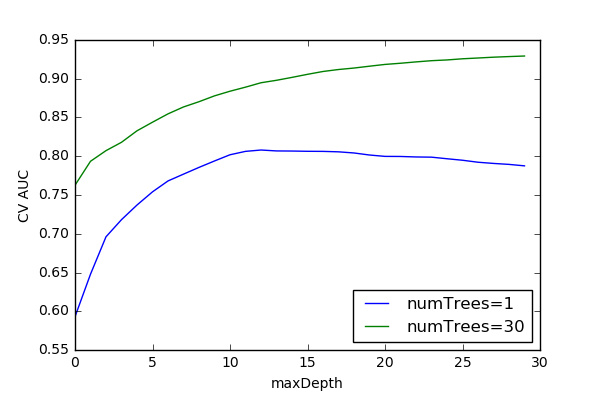
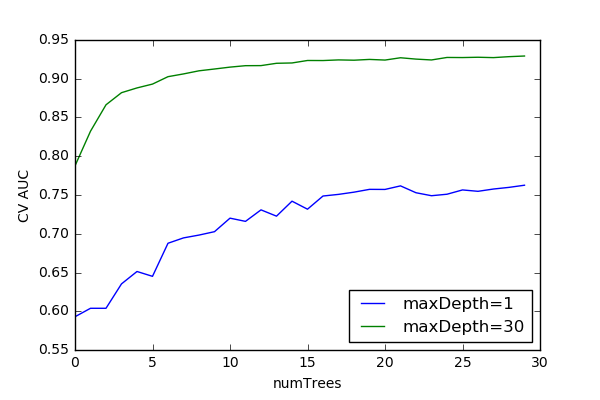
Averaging Ensembles (Bagging)¶
These methods can be used on any model, as they treat the model as a black box. They all train several models and predict using the average (or majority vote) of the ensemble. They reduce variance.
- Pasting. Sample without replacement to train.
- Bagging. Sample with replacement to train.
- Random Subspaces. Use random samples of features to train.
- Random Patches. Bagging+Random Subspaces.
How are these different from Random Forest?
Ensemble Learning with Neural Networks¶
ImageNet Classification with Deep Convolutional Neural Networks¶
Alex Krizhevsky, Ilya Sutskever, Geoffrey E. Hinton
Cited by 126,795
The CNN described in this paper achieves a top-5 error rate of 18.2%. Averaging the predictions of five similar CNNs gives an error rate of 16.4%.
Boosting¶
The goal of boosting is to create a single strong model from a collection of weak models.
The weak models are trained sequentially to strengthen the previously trained model(s).
AdaBoost¶
- Assign uniform weights to data
- Train $model_0$ on data and add to ensemble $M$
- Evaluate data with $M$
- Increase weights of mispredicted data
- Train $model_i$ on re-weighted data and add to ensemble $M$
- Repeat 3-4
The ensemble, $M$, predicts by taking the weighted average of all the weak learners.
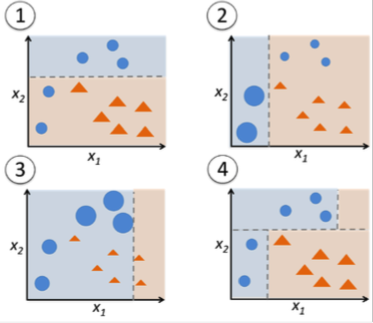
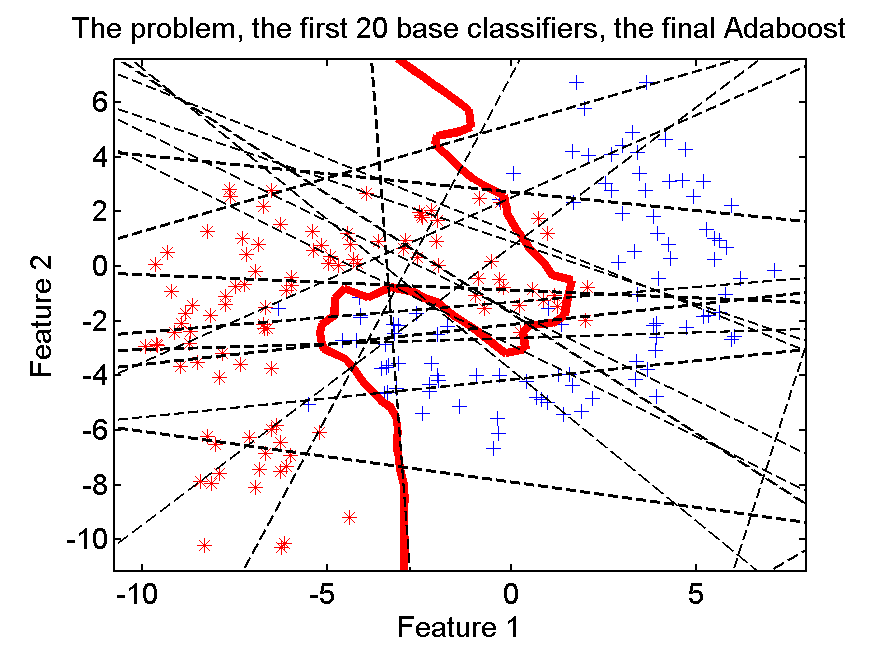

 What are the weak learners?
What are the weak learners?
Can be anything, but often decision stumps, decision trees with only two leaves (max_depth=1).
How are the models weighted?
Proportional to their error
$$\alpha_m = \frac{1}{2}ln\left(\frac{1-\epsilon_m}{\epsilon_m}\right)$$
$\epsilon_m$ is the weighted normalized error for model $m$ (ranges from zero to one)
import matplotlib.pyplot as plt
xvals = np.linspace(0,1,100)
plt.plot(xvals, np.log((1-xvals)/xvals)*0.5)
/tmp/ipykernel_2003701/2736838089.py:3: RuntimeWarning: divide by zero encountered in divide plt.plot(xvals, np.log((1-xvals)/xvals)*0.5) /tmp/ipykernel_2003701/2736838089.py:3: RuntimeWarning: divide by zero encountered in log plt.plot(xvals, np.log((1-xvals)/xvals)*0.5)
[<matplotlib.lines.Line2D at 0x7f3360d2d750>]
%%html
<div id="rfada1x" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#rfada1x';
jQuery(divid).asker({
id: divid,
question: "In a distributed environment, which is faster to train?",
answers: ["Random Forests","AdaBoost","About the same"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
%%html
<div id="rfada2x" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#rfada2x';
jQuery(divid).asker({
id: divid,
question: "In a distributed environment, which is faster to predict?",
answers: ["Random Forests","AdaBoost","About the same"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Gradient Boosted Trees¶
- Train $model_0$ on data and add to ensemble $M$
- Evaluate data with $M$
- Set labels to the residuals ($y - M(x)$)
- Train $model_i$ using new labels
- Optimize multiplier for $model_i$ and add to M
- Repeat 2-5
(Partial) Residuals¶
For a given loss function $L$ we want: $$M_m(x) = M_{m-1}(x) + \left( \min_{model_m} \sum_i^n L\left( y_i, M_{m-1}(x_i) + model_m(x_i) \right) \right)(x)$$ We can view this as gradient descent - we want to "push" $M$'s loss smaller: $$M_m(x) = M_{m-1}(x) - \gamma_m \sum_i^n \nabla_{M_{m-1}} L(y_i, M_{m-1}(x_i))$$ So we want to find a $model_m$ that fits $$ \frac{\partial L(y_i, M_{m-1}(x_i))}{\partial M_{m-1}(x_i)}$$ and then find a $\gamma_m$ that minimizes $$ \sum_i^n L(y_i, M_{m-1} + \gamma_m model_m(x_i))$$
Good News¶
For $L2$ loss $(y-M(x))^2$ you can ignore the previous slide.
Fit $model_m$ to $(y-M_{i-1}(x))$. This is the full residual (and also the gradient of $L$).
Log Loss (Classification)¶
$$L=-\sum_i^N y_{i}\cdot log\left(\frac{1}{1+e^{-M(x_i)}}\right)+(1-y_{i})\cdot log\left(\frac{e^{-M(x_i)}}{1+e^{-M(x_i)}}\right)$$
$${L_i}'=y_{i}-p_{i}$$ where $$p_i = \frac{1}{1+e^{-M(x_i)}}$$
Note: This means we are learning regression trees, not simple classifiers.
Regularization¶
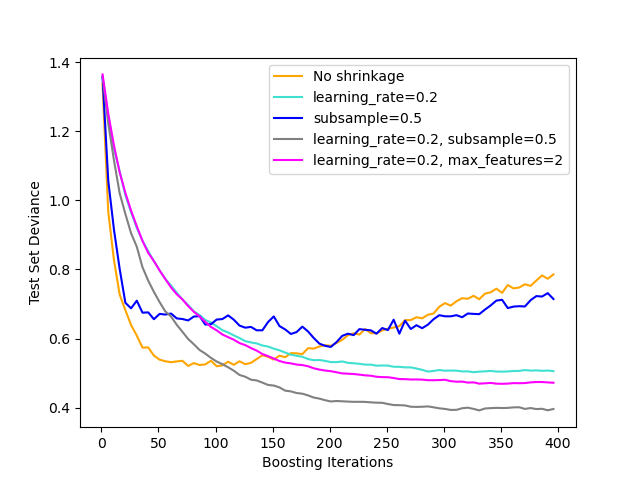
Shrinkage¶
$$M_m(x) = M_{m-1} + v \cdot model_m(x), 0 < v \le 1$$
A learning rate, $v$ can be specified as a regularizer. Each model is prevented from contributing its full amount to the final model. Empirically this results in less overfitting, but at the expense of requiring more iterations (trees).
Stochastic gradient boosting¶
At each iteration train on a sample of the data.
%%html
<div id="adagrad1" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#adagrad1';
jQuery(divid).asker({
id: divid,
question: "In a distributed environment, which is faster to train?",
answers: ["AdaBoost","Gradient Boosted Trees","About the same"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
%%html
<div id="adagrad2" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#adagrad2';
jQuery(divid).asker({
id: divid,
question: "In a distributed environment, which is faster to test?",
answers: ["AdaBoost","Gradient Boosted Trees","About the same"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Gradient Boosting vs. AdaBoost¶
AdaBoost¶
- sequentially updates weight of data ($X$)
- weak learner can be classifier or regressor
- sequential training, parallel testing
- specialization of gradient boosting (exponential loss)
Gradient Boosting¶
- sequentially updates labels ($y$)
- weak learner is regressor
- sequential training, parallel testing
- more general framework than AdaBoost
%%html
<div id="gradnottree" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#gradnottree';
jQuery(divid).asker({
id: divid,
question: "Does gradient boosting only work for decision trees?",
answers: ["Yes","No"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
import matplotlib.pyplot as plt
from sklearn.ensemble import GradientBoostingClassifier
gbt = GradientBoostingClassifier()
gbt.fit(features,labels);
trainerror = gbt.train_score_
plt.plot(trainerror); plt.ylabel("Train Error"); plt.xlabel("Iteration");
Key parameters:
- All decision tree parameters apply (e.g., defaults to max_depth=3)
- n_estimators - number of trees
- n_iter_no_change - early stopping if no change for this many iterations
- learning_rate - amount to discount new learners
- subsample (default 1.0) - stochastic gradient boosting
gbt = GradientBoostingClassifier(learning_rate=1)
gbt.fit(features,labels)
plt.plot(trainerror,label="lr = 0.1")
plt.plot(gbt.train_score_,label="lr = 1.0"); plt.ylabel("Train Error"); plt.xlabel("Iteration");
plt.legend();
%%html
<div id="gbq" style="width: 500px"></div>
<script>
var divid = '#gbq';
jQuery(divid).asker({
id: divid,
question: "What to you expect to happen to the CV performance of GBT as the number of iterations is increased?",
answers: ["Go up","Go down","Go up then down","Go up then level off"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import StratifiedKFold
skf = StratifiedKFold(3)
train,test = next(skf.split(features,labels))
rftrainaucs = []
rftestaucs = []
for d in range(1,200):
rf = RandomForestClassifier(n_estimators=d,max_depth=2,n_jobs=-1)
rf.fit(features[train],labels[train])
rftrainaucs.append(roc_auc_score(labels[train],rf.predict_proba(features[train])[:,1]))
rftestaucs.append(roc_auc_score(labels[test],rf.predict_proba(features[test])[:,1]))
gbttrainaucs = []
gbttestaucs = []
gbt = GradientBoostingClassifier(n_estimators=1,max_depth=2,warm_start=True,learning_rate=.9)
for d in range(1,200):
gbt.n_estimators = d
gbt.fit(features[train],labels[train])
gbttrainaucs.append(roc_auc_score(labels[train],gbt.predict_proba(features[train])[:,1]))
gbttestaucs.append(roc_auc_score(labels[test],gbt.predict_proba(features[test])[:,1]))
plt.figure(figsize=(8,4),dpi=100)
plt.plot(rftrainaucs,'b--',label='RF train'); plt.plot(rftestaucs,'b-',label='RF test')
plt.plot(gbttrainaucs,'g--',label='GBT train'); plt.plot(gbttestaucs,'g-',label='GBT test')
plt.legend();
Boosting Resists Overfitting¶
This is an empirical observation (and resists isn't the same as avoids). Some intuitions why:
- each new iteration
- has less effect (diluted)
- is localized (to mispredicted examples)
- stagewise estimation slows learning process (parameters aren't fit simultaneously)
- margin theory? https://www.cc.gatech.edu/~isbell/tutorials/boostingmargins.pdf
- narrow decision boundaries for "tricky" examples

Case Study: XGBoost¶
https://arxiv.org/abs/1603.02754

The impact of the system has been widely recognized in a number of machine learning and data mining challenges. Take the challenges hosted by the machine learning competition site Kaggle for example. Among the 29 challenge winning solutions 3 published at Kaggle's blog during 2015, 17 solutions used XGBoost. Among these solutions, eight solely used XGBoost to train the model, while most others combined XGBoost with neural nets in ensembles. For comparison, the second most popular method, deep neural nets, was used in 11 solutions. The success of the system was also witnessed in KDDCup 2015, where XGBoost was used by every winning team in the top-10. Moreover, the winning teams reported that ensemble methods outperform a well-con gured XGBoost by only a small amount.
XGBoost¶
- a novel distributed weighted quantile sketch algorithm
- sparsity aware split finding
- column-based memory layout
- cache optimization
- I/O aware system design
Distributed weighted quantile sketch¶
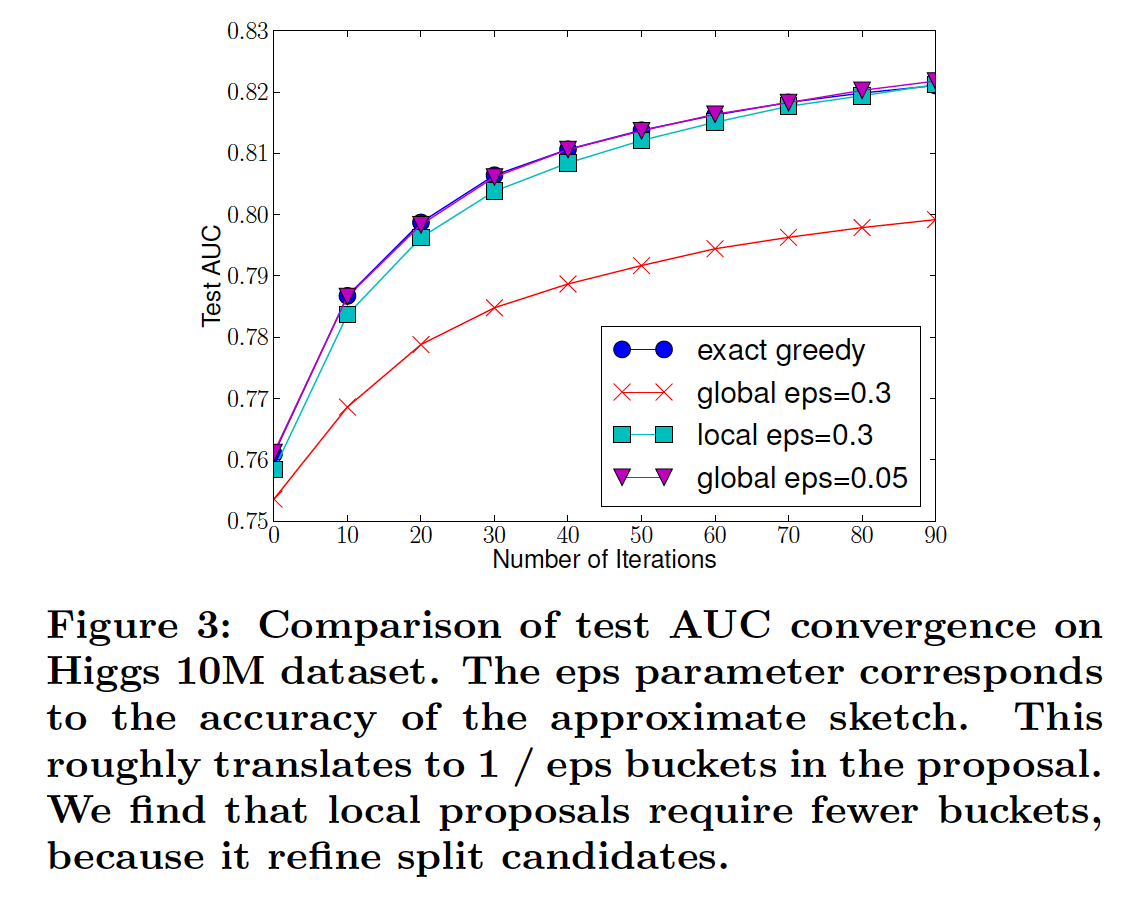
Spark: Subsamples data at start to compute a histogram and determine a fixed number of reasonable split points (global split).
XGBoost: Compute summary statistics (quantile sketch) with theoretical bounds on quality to determine split points. Can do once at initialization of tree construction (global) or at every split in the tree (local).
Split points are both based on the distribution of the feature value and the associated label.
%%html
<div id="boostlg" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#boostlg';
jQuery(divid).asker({
id: divid,
question: "If decision stumps are being used, which splitting policy would you expect to do better?",
answers: ["global","local","the same"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Sparsity¶
 |  |
Column-based layout¶
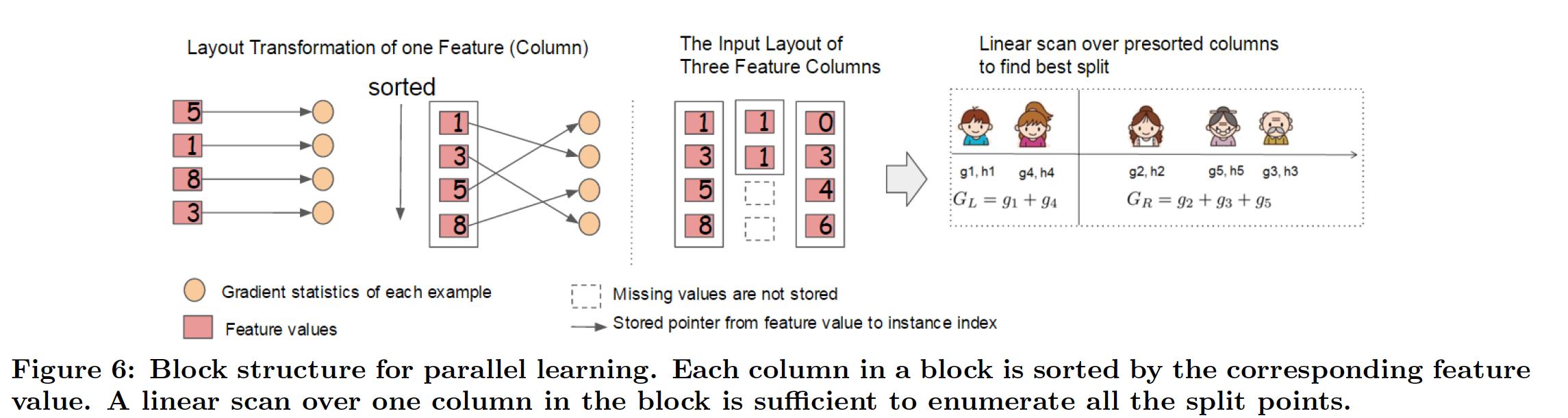
Cache Optimization¶
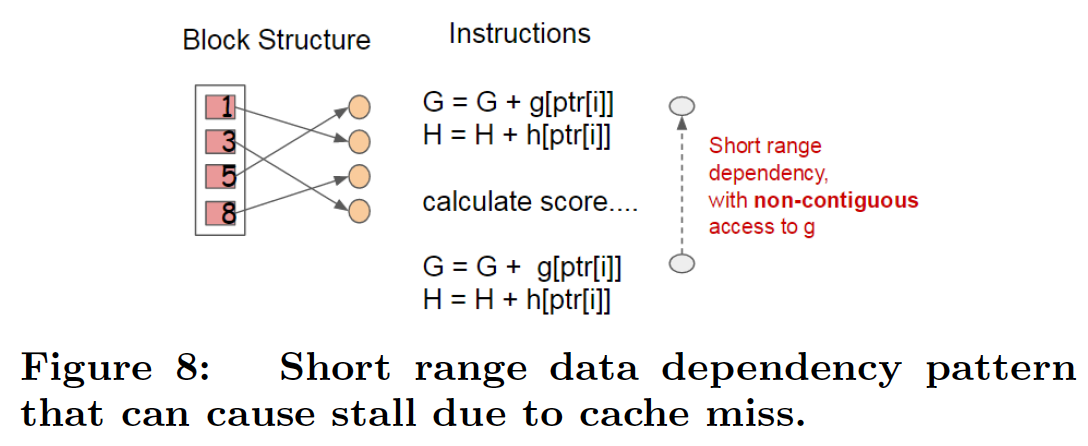
int i;
const Entry *it;
const int align_step = d_step * kBuffer;
// internal cached loop
for (it = begin; it != align_end; it += align_step) {
const Entry *p;
for (i = 0, p = it; i < kBuffer; ++i, p += d_step) {
buf_position[i] = position_[p->index];
buf_gpair[i] = gpair[p->index];
}
for (i = 0, p = it; i < kBuffer; ++i, p += d_step) {
const int nid = buf_position[i];
if (nid < 0 || !interaction_constraints_.Query(nid, fid)) { continue; }
this->UpdateEnumeration(nid, buf_gpair[i],
p->fvalue, d_step,
fid, c, temp, evaluator);
}
}
Cache Optimization¶
I/O Aware System Design¶
Data is grouped into blocks
 Data of next block is prefetched concurrently while current block is processed
Data of next block is prefetched concurrently while current block is processed- Blocks are compressed
- Blocks are striped across multiple disks if available

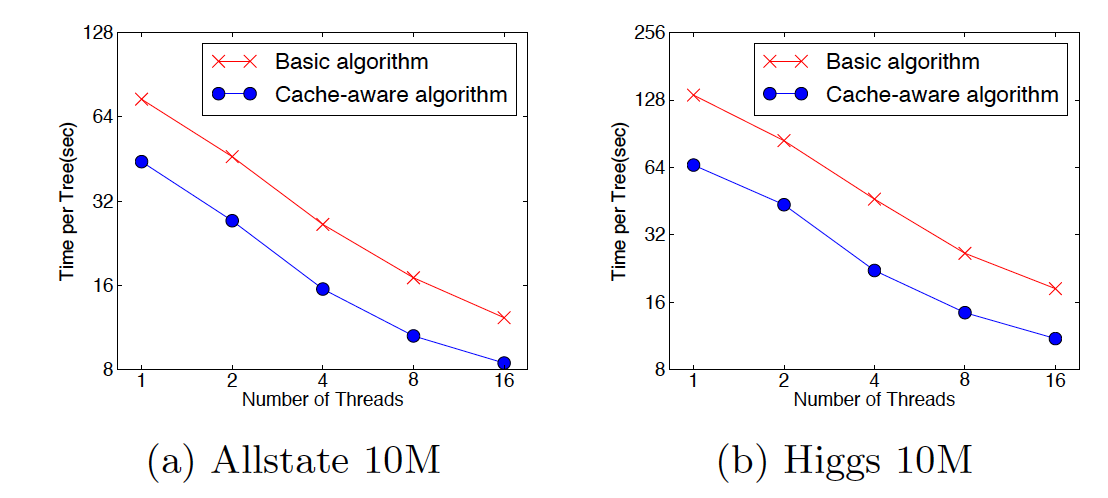
XGBoost Overall Performance¶
 |  |
XGBoost Scalability¶
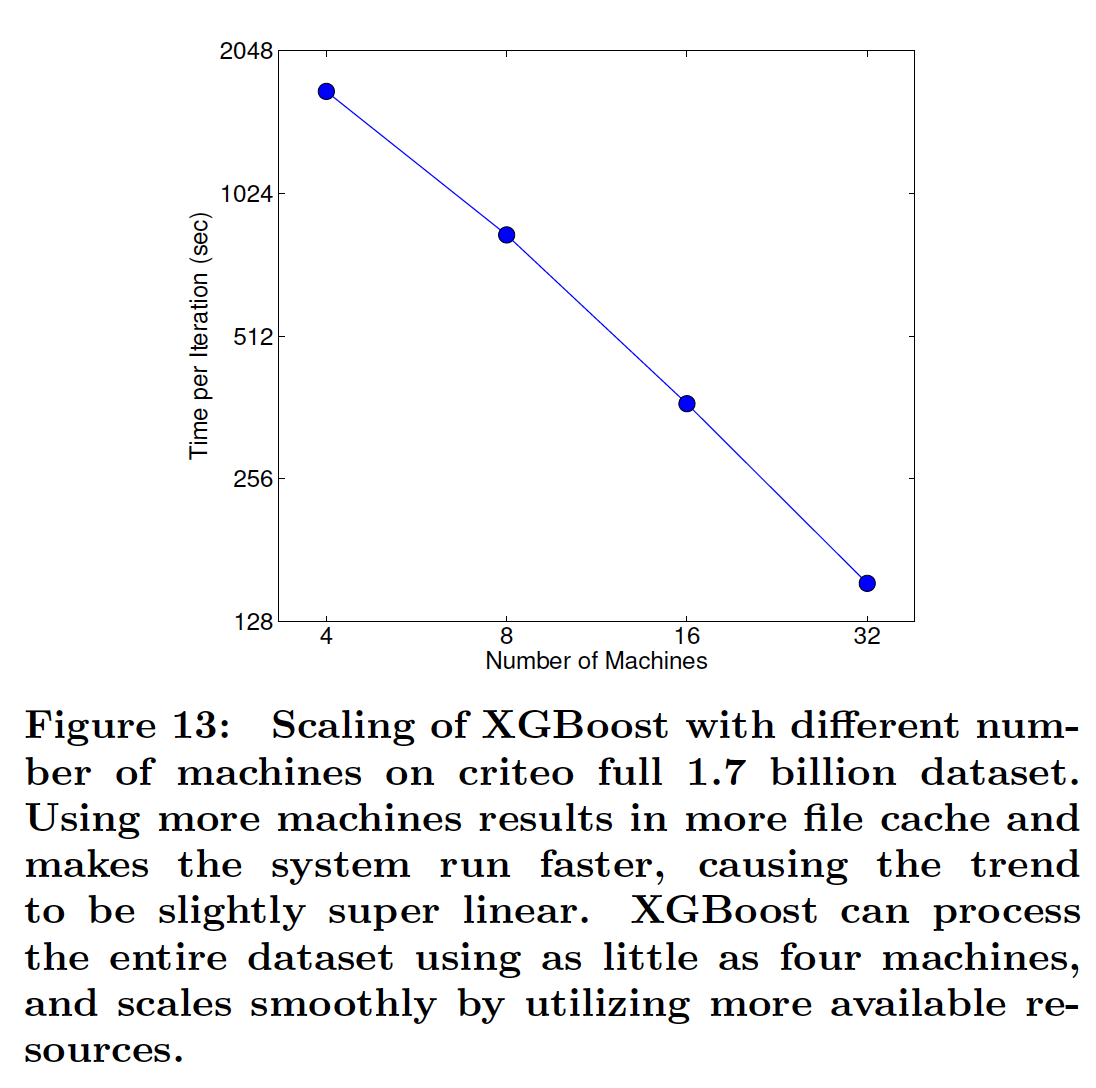
import xgboost as xgb
param = {'max_depth':3,'eta':0.9,'objective':'binary:logistic','eval_metric':'logloss','tree_method':'exact'}
trainset = xgb.DMatrix(features[train],label=labels[train])
xtrainaucs = []
xtestaucs = []
xgbt = None
for d in range(1,200):
xgbt = xgb.train(param,trainset,1,xgb_model=xgbt)
xtrainaucs.append(roc_auc_score(labels[train],xgbt.predict(xgb.DMatrix(features[train]))))
xtestaucs.append(roc_auc_score(labels[test],xgbt.predict(xgb.DMatrix(features[test]))))
plt.figure(figsize=(8,4),dpi=100)
plt.plot(rftrainaucs,'b--',label='RF train'); plt.plot(rftestaucs,'b-',label='RF test')
plt.plot(gbttrainaucs,'g--',label='GBT train'); plt.plot(gbttestaucs,'g-',label='GBT test')
plt.plot(xtrainaucs,'r--',label='XGB train'); plt.plot(xtestaucs,'r-',label='XGB test')
plt.legend();
%%time
rf = RandomForestClassifier(n_estimators=200,max_depth=2,n_jobs=-1)
rf.fit(features[train],labels[train]);
CPU times: user 1.82 s, sys: 43.8 ms, total: 1.87 s Wall time: 531 ms
%%time
gbt = GradientBoostingClassifier(n_estimators=200,max_depth=2,learning_rate=.9)
gbt.fit(features[train],labels[train]);
CPU times: user 1min, sys: 59.6 ms, total: 1min Wall time: 1min
%%time
xgbt = xgb.train(param,trainset,200)
CPU times: user 2min 11s, sys: 113 ms, total: 2min 11s Wall time: 5.72 s
%%time
treeparms = {'tree_method':'hist','max_depth':2,'eta':0.9,'objective':'binary:logistic','eval_metric':'logloss'}
xgbt = xgb.train(treeparms,trainset,200)
CPU times: user 25.6 s, sys: 39.8 ms, total: 25.7 s Wall time: 1.12 s
%%time
gpuparms = {'tree_method':'gpu_hist','max_depth':2,'eta':0.9,'objective':'binary:logistic','eval_metric':'logloss'}
xgbt = xgb.train(gpuparms,trainset,200)
CPU times: user 510 ms, sys: 32.3 ms, total: 542 ms Wall time: 293 ms
gpuxtrainaucs = []
gpuxtestaucs = []
xgbt = None
for d in range(1,200):
xgbt = xgb.train(gpuparms,trainset,1,xgb_model=xgbt)
gpuxtrainaucs.append(roc_auc_score(labels[train],xgbt.predict(xgb.DMatrix(features[train]))))
gpuxtestaucs.append(roc_auc_score(labels[test],xgbt.predict(xgb.DMatrix(features[test]))))
plt.figure(figsize=(8,4),dpi=100)
plt.plot(xtrainaucs,'r--',label='XGB train'); plt.plot(xtestaucs,'r-',label='XGB test')
plt.plot(gpuxtrainaucs,'m--',label='XGB GPU train'); plt.plot(gpuxtestaucs,'m-',label='XGB GPU test')
plt.legend();
They are identical, despite the histogram approximation.
Why?
def plot_gpt(X,Y,depth=1):
gbt = GradientBoostingClassifier(max_depth=depth,n_estimators=7,learning_rate=1).fit(X,Y)
fig, axes = plt.subplots(ncols=4,nrows=2,figsize=(16,8))
cm = plt.cm.PiYG
for i in range(8):
ax = axes[i//4][i%4]
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.01), np.arange(y_min, y_max, 0.01))
ax.title
if i < 7: #individual trees
Z = gbt.estimators_[i][0].predict(np.c_[xx.ravel(), yy.ravel()])
else: #full predictions
Z = gbt.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
ax.contourf(xx, yy, Z, cmap=cm, alpha=.5)
ax.scatter(X[:,0],X[:,1],c=Y,edgecolors='k',cmap=cm)
from sklearn.datasets import make_moons, make_circles, make_classification
X,Y = make_moons(n_samples=100,noise=.1,random_state=3)
plot_gpt(X,Y)
X,Y = make_circles(n_samples=100,noise=.1,random_state=3)
plot_gpt(X,Y)
X,Y = make_classification(n_samples=100, n_features=2, n_redundant=0,
n_informative=2, random_state=2,
n_clusters_per_class=1)
plot_gpt(X,Y)
X = np.array([[-1,-1],[-1,1],[1,-1],[1,1]])
Y = np.array([1,0,0,1])
plt.figure(figsize=(4,4))
plt.scatter(X[:,0],X[:,1],c=Y,edgecolors='k',cmap=plt.cm.PiYG)
plt.ylim(-1.5,1.5); plt.xlim(-1.5,1.5)
(-1.5, 1.5)
plot_gpt(X,Y)
%%html
<div id="xorq" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#xorq';
jQuery(divid).asker({
id: divid,
question: "What will happen if we increase the tree depth?",
answers: ["Perfect fit","Same problem","Better, not perfect"],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
plot_gpt(X,Y,depth=2)