 Molecular visualization is an essential tool in biology and chemistry.
3Dmol is a JavaScript library for molecular visualization
that uses WebGL to provide hardware accelerated 3D graphics.
Molecular visualization is an essential tool in biology and chemistry.
3Dmol is a JavaScript library for molecular visualization
that uses WebGL to provide hardware accelerated 3D graphics.
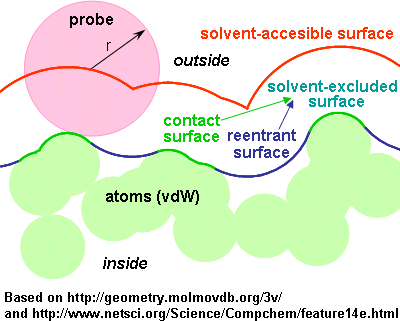
In this project you will optimize molecular surface generation code. The code takes a list of atoms and computes a 3D mesh representing the solvent excluded surface using the EDTSurf algorithm.
A reference JavaScript implementation is provided on this page and is available through github. Note that the X-Large example below does not work since WebGL buffers are limited to 65,536 elements. For the purposes of the project, you do not need to support meshes larger than 64k, however if you are splitting up the mesh to parallelize it, it should be straightfoward to get larger meshes working.
Please contact me (dkoes@pitt.edu) if you have any questions. You have two options in tackling this project.
In this option the surface generation code runs exclusively on the client using JavaScript and standard APIs. You may choose to optimize for any widely available browser. For example, you might use asm.js and target Firefox. You may not rely on plugins, Java, or Flash because these are evil incarnate. The exception is that you may use WebCL (since it is a standard, albeit a poorly supported one) even if this means using the Nokia plugin or Chromium-WebCL.
In this option you may implement the surface generation code in any language and run it on a server. You are strongly encouraged to use CUDA, and access to workstation(s) with nVidia GTX 980 graphics cards can be provided if needed. The EDTSurf code will serve as the reference implementation in this case.