%%html
<script src="https://bits.csb.pitt.edu/preamble.js"></script>
Reminder¶
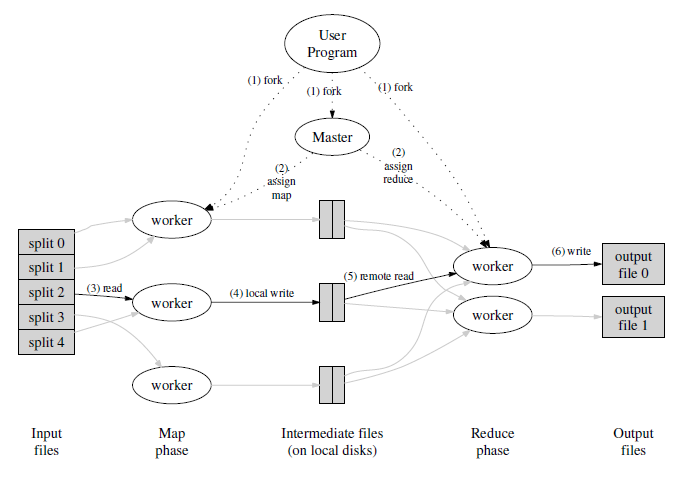
Hadoop¶

Hadoop¶
Hadoop is a framework of open-source modules for scalable, fault-tolerant distributed computing developed by the Apache Foundation. It is named after a developer's child's toy elephant.
- Hadoop Distributed File System (HDFS): A distributed file system that provides high-throughput access to application data.
- Hadoop MapReduce: A YARN-based system for parallel processing of large data sets.
HDFS¶
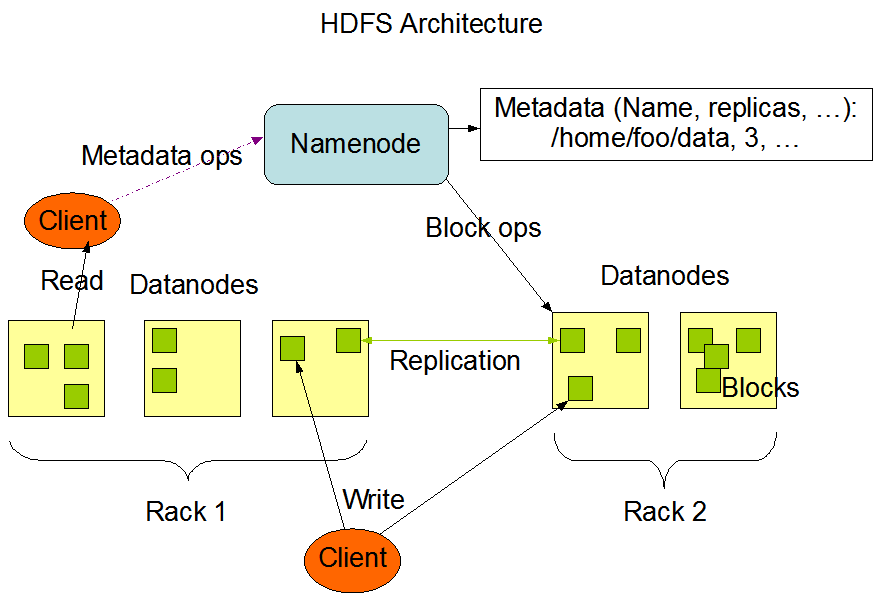
HDFS Design Goals¶
 Tolerate hardware failure
Tolerate hardware failure- Optimize for streaming data access - emphasize throughput over latency
- Optimize for large data sets
- Simple coherency model (write once, read many)
- "Moving Computation is Cheaper than Moving Data"
- Portability
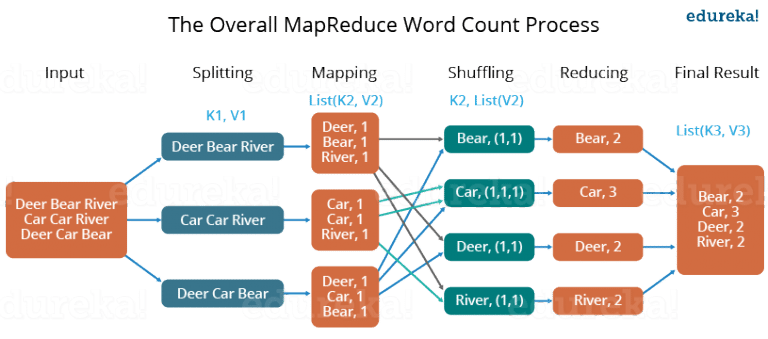
MapReduce¶
The input is a list of $(k_1,v_1)$ key-value pairs.
The programmer specifies:
a map function: $map(k_1,v_1) \rightarrow list(k_2,v_2)$
that performs some computation on a single key-value pairs and generates new key value pairs (potentially with very different keys and values).
a reduce function: $reduce(k_2,list(v_2)) \rightarrow list(v_3)$
that is provided with the sorted list of values for a given key.
Partitioning¶
MapReduce depends on being able to partition the intermediate (and final) values based on the intermediate key into independent partitions.
This means it needs to be able to map any distribution of keys into a roughly uniform grouping.
To do this it uses hashing: $hash(k_2) \mod R$
Properties of a hash function:
- always returns a number for an object
- two equal objects will always have the same number
- two different objects will not always have the same number
A good hash function:
- depends on every bit of the input
- generates uniformly distributed values
- is fast

Zaharia, Matei, et al. "Spark: cluster computing with working sets." HotCloud 10 (2010): 10-10. http://static.usenix.org/legacy/events/hotcloud10/tech/full_papers/Zaharia.pdf
Zaharia, Matei, et al. "Resilient distributed datasets: A fault-tolerant abstraction for in-memory cluster computing." Proceedings of the 9th USENIX conference on Networked Systems Design and Implementation. USENIX Association, 2012. https://www.usenix.org/conference/nsdi12/technical-sessions/presentation/zaharia
Goals of Spark¶
- Support iterative algorithms (significant reuse of data)
- Support interactive analysis (low latency startup)
- High performance
- Fault tolerance
- Clean programming abstraction
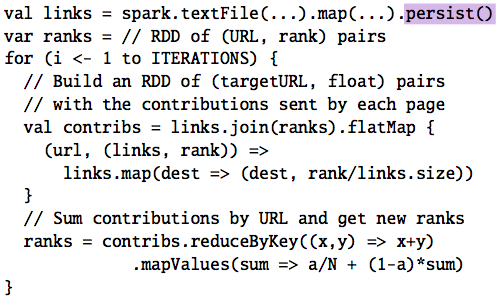
Key idea: Resilient Distributed Datasets
Resilient Distributed Datasets¶
A distributed memory abstraction
Programming abstraction¶
- read-only set of (key,value) records
- or just a list of values
- can only be created by
- loading data (e.g. HDFS file)
- as a result of a transformation of another RDD
- actions convert RDDs to regular files or other output
Resilient Distributed Datasets¶
A distributed memory abstraction
Implementation¶
- data is stored in memory (usually)
- can request data be stored on disk
- transformations are lazy - data isn't generated until it is needed by an action
- every RDD knows how it was created through its lineage
- persistent RDDs stay in memory - don't need to be recomputed
- will spill to disk if necessary
- every RDD has a partition specifying what nodes the data is stored on
- not replicated
Transformations¶
An RDD transformation creates a new RDD from an existing one.
Both map and reduceByKey are transformations, but many more are supported:
http://spark.apache.org/docs/latest/rdd-programming-guide.html#transformations
- map(func)
- filter(func)
- flatMap(func)
- mapPartitions(func)
- mapPartitionsWithIndex(func)
- sample(withReplacement, fraction, seed)
- union(otherDataset)
- intersection(otherDataset)
- distinct([numTasks]))
- groupByKey([numTasks])
- reduceByKey(func, [numTasks])
- aggregateByKey(zeroValue)(seqOp, combOp, [numTasks])
- sortByKey([ascending], [numTasks])
- join(otherDataset, [numTasks])
- cogroup(otherDataset, [numTasks])
- cartesian(otherDataset)
- pipe(command, [envVars])
- coalesce(numPartitions)
- repartition(numPartitions)
- repartitionAndSortWithinPartitions(partitioner)
Actions¶
Actions work on an RDD to produce a non-RDD result accessible to the driver program
http://spark.apache.org/docs/latest/rdd-programming-guide.html#actions
- reduce(func)
- collect()
- count()
- first()
- take(n)
- takeSample(withReplacement, num, [seed])
- takeOrdered(n, [ordering])
- saveAsTextFile(path)
- foreach(func)
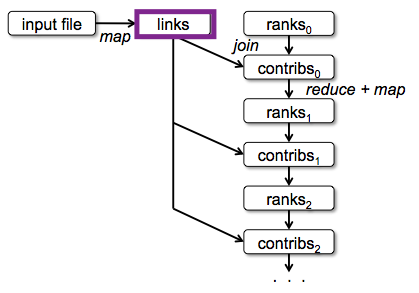
Lineage¶
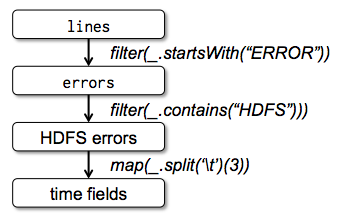 Every RDD (here "lines","errors", "HDFS errors", and "time fields") knows how it was computed.
Every RDD (here "lines","errors", "HDFS errors", and "time fields") knows how it was computed.
Dependencies¶
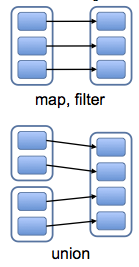
Narrow dependencies: Each partition of the parent RDD is used by at most one partition of the child.
- can be resolved without any inter-node communication
Dependencies¶
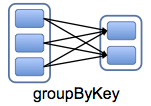
Wide dependencies: Each partition of the parent RDD is used by (potentially) more than one partition of the child.
- Require inter-node communication (Unles you are lucky)
 http://horicky.blogspot.com/2015/02/big-data-processing-in-spark.html
http://horicky.blogspot.com/2015/02/big-data-processing-in-spark.html
Scheduling¶
Narrow dependencies are grouped into stages. The tasks of each stage are executed on the same node. A stage is scheduled once all of the stages it is dependent on are available (but see https://issues.apache.org/jira/browse/SPARK-2387).
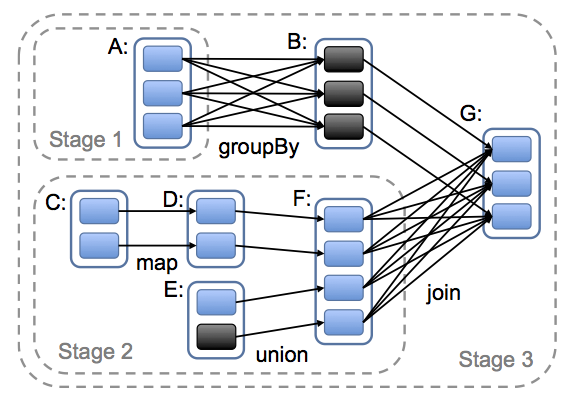
Black boxes are partitions that are already in memory (thanks to persist).
Delay Scheduling (Tasks)¶
Spark will attempt to assign a task (e.g. map on a partition) to the node that has the data.
If the node is busy with other tasks, it will wait a small amount of time.
If the node is still busy and there is a free node elsewhere, it will run on that node, loading the data across the network.
User Interfaces¶
High-Level
- Arrays: Parallel NumPy
- DataFrames: Parallel Pandas
- Bags: Parallel lists (pySpark)
- Machine Learning: Parallel Scikit-Learn
Low-Level
- Delayed: Lazy parallel function evaluation
- Futures: Real-time parallel function evaluation
import dask.array as da
x = da.random.uniform(low=0, high=10, size=(10000, 10000), # normal numpy code
chunks=(5000, 5000)) # break into chunks of size 5000x5000
y = x + x.T - x.mean(axis=0) # Use normal syntax for high level algorithms
x
|
y
|
y.visualize()
We have not computed anything yet!¶
result = y.compute()
type(result)
numpy.ndarray
x = da.random.uniform(low=0, high=10, size=(10000, 10000), # normal numpy code
chunks=(5000, 5000)) # break into chunks of size 5000x5000
y = x + x.T - x.mean(axis=0)
y
|
%%timeit
y.compute()
467 ms ± 6.04 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x + x.T - x.mean(axis=0)
y
|
%timeit y.compute()
345 ms ± 20.5 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(10000, 10000))
y = x + x.T - x.mean(axis=0)
y
|
%timeit y.compute()
1.17 s ± 7.91 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
y.visualize()
import numpy as np
%%timeit
x = np.random.uniform(low=0, high=10, size=(10000, 10000))
y = x + x.T - x.mean(axis=0)
1.15 s ± 8.3 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
import dask.dataframe as dd
df = dd.read_csv('Hospital_Inpatient_Discharges__SPARCS_De-Identified___2014.csv')
This file is 908MB. Be default will create 64MB chunks.
df
| Health Service Area | Hospital County | Operating Certificate Number | Facility ID | Facility Name | Age Group | Zip Code - 3 digits | Gender | Race | Ethnicity | Length of Stay | Type of Admission | Patient Disposition | Discharge Year | CCS Diagnosis Code | CCS Diagnosis Description | CCS Procedure Code | CCS Procedure Description | APR DRG Code | APR DRG Description | APR MDC Code | APR MDC Description | APR Severity of Illness Code | APR Severity of Illness Description | APR Risk of Mortality | APR Medical Surgical Description | Payment Typology 1 | Payment Typology 2 | Payment Typology 3 | Attending Provider License Number | Operating Provider License Number | Other Provider License Number | Birth Weight | Abortion Edit Indicator | Emergency Department Indicator | Total Charges | Total Costs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| npartitions=14 | |||||||||||||||||||||||||||||||||||||
| object | object | int64 | int64 | object | object | int64 | object | object | object | int64 | object | object | int64 | int64 | object | int64 | object | int64 | object | int64 | object | int64 | object | object | object | object | object | float64 | int64 | float64 | float64 | int64 | object | object | object | object | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
df.head()
/Library/Python/3.9/site-packages/dask/dataframe/io/csv.py:181: DtypeWarning: Columns (10) have mixed types. Specify dtype option on import or set low_memory=False. df = reader(bio, **kwargs)
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) /var/folders/c_/pwm7n7_174724g8zkkqlpr3m0000gn/T/ipykernel_84781/964094849.py in <module> ----> 1 df.head() /Library/Python/3.9/site-packages/dask/dataframe/core.py in head(self, n, npartitions, compute) 1138 # No need to warn if we're already looking at all partitions 1139 safe = npartitions != self.npartitions -> 1140 return self._head(n=n, npartitions=npartitions, compute=compute, safe=safe) 1141 1142 def _head(self, n, npartitions, compute, safe): /Library/Python/3.9/site-packages/dask/dataframe/core.py in _head(self, n, npartitions, compute, safe) 1172 1173 if compute: -> 1174 result = result.compute() 1175 return result 1176 /Library/Python/3.9/site-packages/dask/base.py in compute(self, **kwargs) 286 dask.base.compute 287 """ --> 288 (result,) = compute(self, traverse=False, **kwargs) 289 return result 290 /Library/Python/3.9/site-packages/dask/base.py in compute(traverse, optimize_graph, scheduler, get, *args, **kwargs) 569 postcomputes.append(x.__dask_postcompute__()) 570 --> 571 results = schedule(dsk, keys, **kwargs) 572 return repack([f(r, *a) for r, (f, a) in zip(results, postcomputes)]) 573 /Library/Python/3.9/site-packages/dask/threaded.py in get(dsk, result, cache, num_workers, pool, **kwargs) 77 pool = MultiprocessingPoolExecutor(pool) 78 ---> 79 results = get_async( 80 pool.submit, 81 pool._max_workers, /Library/Python/3.9/site-packages/dask/local.py in get_async(submit, num_workers, dsk, result, cache, get_id, rerun_exceptions_locally, pack_exception, raise_exception, callbacks, dumps, loads, chunksize, **kwargs) 505 _execute_task(task, data) # Re-execute locally 506 else: --> 507 raise_exception(exc, tb) 508 res, worker_id = loads(res_info) 509 state["cache"][key] = res /Library/Python/3.9/site-packages/dask/local.py in reraise(exc, tb) 313 if exc.__traceback__ is not tb: 314 raise exc.with_traceback(tb) --> 315 raise exc 316 317 /Library/Python/3.9/site-packages/dask/local.py in execute_task(key, task_info, dumps, loads, get_id, pack_exception) 218 try: 219 task, data = loads(task_info) --> 220 result = _execute_task(task, data) 221 id = get_id() 222 result = dumps((result, id)) /Library/Python/3.9/site-packages/dask/core.py in _execute_task(arg, cache, dsk) 117 # temporaries by their reference count and can execute certain 118 # operations in-place. --> 119 return func(*(_execute_task(a, cache) for a in args)) 120 elif not ishashable(arg): 121 return arg /Library/Python/3.9/site-packages/dask/optimization.py in __call__(self, *args) 967 if not len(args) == len(self.inkeys): 968 raise ValueError("Expected %d args, got %d" % (len(self.inkeys), len(args))) --> 969 return core.get(self.dsk, self.outkey, dict(zip(self.inkeys, args))) 970 971 def __reduce__(self): /Library/Python/3.9/site-packages/dask/core.py in get(dsk, out, cache) 147 for key in toposort(dsk): 148 task = dsk[key] --> 149 result = _execute_task(task, cache) 150 cache[key] = result 151 result = _execute_task(out, cache) /Library/Python/3.9/site-packages/dask/core.py in _execute_task(arg, cache, dsk) 117 # temporaries by their reference count and can execute certain 118 # operations in-place. --> 119 return func(*(_execute_task(a, cache) for a in args)) 120 elif not ishashable(arg): 121 return arg /Library/Python/3.9/site-packages/dask/dataframe/io/csv.py in __call__(self, part) 126 127 # Call `pandas_read_text` --> 128 df = pandas_read_text( 129 self.reader, 130 block, /Library/Python/3.9/site-packages/dask/dataframe/io/csv.py in pandas_read_text(reader, b, header, kwargs, dtypes, columns, write_header, enforce, path) 181 df = reader(bio, **kwargs) 182 if dtypes: --> 183 coerce_dtypes(df, dtypes) 184 185 if enforce and columns and (list(df.columns) != list(columns)): /Library/Python/3.9/site-packages/dask/dataframe/io/csv.py in coerce_dtypes(df, dtypes) 282 rule.join(filter(None, [dtype_msg, date_msg])) 283 ) --> 284 raise ValueError(msg) 285 286 ValueError: Mismatched dtypes found in `pd.read_csv`/`pd.read_table`. +-----------------------------------+---------+----------+ | Column | Found | Expected | +-----------------------------------+---------+----------+ | Attending Provider License Number | float64 | int64 | | Facility ID | float64 | int64 | | Length of Stay | object | int64 | | Operating Certificate Number | float64 | int64 | | Payment Typology 3 | object | float64 | | Zip Code - 3 digits | object | int64 | +-----------------------------------+---------+----------+ The following columns also raised exceptions on conversion: - Length of Stay ValueError("invalid literal for int() with base 10: '120 +'") - Payment Typology 3 ValueError("could not convert string to float: 'Private Health Insurance'") - Zip Code - 3 digits ValueError("invalid literal for int() with base 10: 'OOS'") Usually this is due to dask's dtype inference failing, and *may* be fixed by specifying dtypes manually by adding: dtype={'Attending Provider License Number': 'float64', 'Facility ID': 'float64', 'Length of Stay': 'object', 'Operating Certificate Number': 'float64', 'Payment Typology 3': 'object', 'Zip Code - 3 digits': 'object'} to the call to `read_csv`/`read_table`.
import pandas as pd
from numpy import dtype
dtypes= {'Health Service Area': dtype('O'),
'Hospital County': dtype('O'),
'Operating Certificate Number': dtype('float64'),
'Facility ID': dtype('float64'),
'Facility Name': dtype('O'),
'Age Group': dtype('O'),
'Zip Code - 3 digits': dtype('O'),
'Gender': dtype('O'),
'Race': dtype('O'),
'Ethnicity': dtype('O'),
'Length of Stay': dtype('O'),
'Type of Admission': dtype('O'),
'Patient Disposition': dtype('O'),
'Discharge Year': dtype('int64'),
'CCS Diagnosis Code': dtype('int64'),
'CCS Diagnosis Description': dtype('O'),
'CCS Procedure Code': dtype('int64'),
'CCS Procedure Description': dtype('O'),
'APR DRG Code': dtype('int64'),
'APR DRG Description': dtype('O'),
'APR MDC Code': dtype('int64'),
'APR MDC Description': dtype('O'),
'APR Severity of Illness Code': dtype('int64'),
'APR Severity of Illness Description': dtype('O'),
'APR Risk of Mortality': dtype('O'),
'APR Medical Surgical Description': dtype('O'),
'Payment Typology 1': dtype('O'),
'Payment Typology 2': dtype('O'),
'Payment Typology 3': dtype('O'),
'Attending Provider License Number': dtype('float64'),
'Operating Provider License Number': dtype('float64'),
'Other Provider License Number': dtype('float64'),
'Birth Weight': dtype('int64'),
'Abortion Edit Indicator': dtype('O'),
'Emergency Department Indicator': dtype('O'),
'Total Charges': dtype('O'),
'Total Costs': dtype('O')}
df = dd.read_csv('Hospital_Inpatient_Discharges__SPARCS_De-Identified___2014.csv',dtype=dtypes)
df.head()
| Health Service Area | Hospital County | Operating Certificate Number | Facility ID | Facility Name | Age Group | Zip Code - 3 digits | Gender | Race | Ethnicity | ... | Payment Typology 2 | Payment Typology 3 | Attending Provider License Number | Operating Provider License Number | Other Provider License Number | Birth Weight | Abortion Edit Indicator | Emergency Department Indicator | Total Charges | Total Costs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Western NY | Allegany | 226700.0 | 37.0 | Cuba Memorial Hospital Inc | 30 to 49 | 147 | F | White | Not Span/Hispanic | ... | NaN | NaN | 90335341.0 | NaN | NaN | 0 | N | Y | $9546.85 | $10998.39 |
| 1 | Western NY | Allegany | 226700.0 | 37.0 | Cuba Memorial Hospital Inc | 50 to 69 | 147 | F | White | Not Span/Hispanic | ... | NaN | NaN | 90335341.0 | NaN | NaN | 0 | N | Y | $11462.75 | $10275.43 |
| 2 | Western NY | Allegany | 226700.0 | 37.0 | Cuba Memorial Hospital Inc | 18 to 29 | 147 | M | White | Not Span/Hispanic | ... | NaN | NaN | 90335341.0 | 167816.0 | NaN | 0 | N | Y | $1609.40 | $2285.55 |
| 3 | Western NY | Allegany | 226700.0 | 37.0 | Cuba Memorial Hospital Inc | 18 to 29 | 147 | F | White | Not Span/Hispanic | ... | NaN | NaN | 90335341.0 | 167816.0 | NaN | 0 | N | Y | $2638.75 | $3657.95 |
| 4 | Western NY | Allegany | 226700.0 | 37.0 | Cuba Memorial Hospital Inc | 18 to 29 | 147 | F | White | Not Span/Hispanic | ... | NaN | NaN | 90335341.0 | NaN | NaN | 0 | N | Y | $3538.25 | $4069.85 |
5 rows × 37 columns
dfsingle = dd.read_csv('Hospital_Inpatient_Discharges__SPARCS_De-Identified___2014.csv',blocksize=None,dtype=dtypes)
dfsingle
| Health Service Area | Hospital County | Operating Certificate Number | Facility ID | Facility Name | Age Group | Zip Code - 3 digits | Gender | Race | Ethnicity | Length of Stay | Type of Admission | Patient Disposition | Discharge Year | CCS Diagnosis Code | CCS Diagnosis Description | CCS Procedure Code | CCS Procedure Description | APR DRG Code | APR DRG Description | APR MDC Code | APR MDC Description | APR Severity of Illness Code | APR Severity of Illness Description | APR Risk of Mortality | APR Medical Surgical Description | Payment Typology 1 | Payment Typology 2 | Payment Typology 3 | Attending Provider License Number | Operating Provider License Number | Other Provider License Number | Birth Weight | Abortion Edit Indicator | Emergency Department Indicator | Total Charges | Total Costs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| npartitions=1 | |||||||||||||||||||||||||||||||||||||
| object | object | float64 | float64 | object | object | object | object | object | object | object | object | object | int64 | int64 | object | int64 | object | int64 | object | int64 | object | int64 | object | object | object | object | object | object | float64 | float64 | float64 | int64 | object | object | object | object | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
df['Gender'].value_counts()
Dask Series Structure:
npartitions=1
int64
...
Name: Gender, dtype: int64
Dask Name: value-counts-agg, 45 tasks
%%timeit
df['Gender'].value_counts().compute()
6.8 s ± 157 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
dfsingle['Gender'].value_counts()
Dask Series Structure:
npartitions=1
int64
...
Name: Gender, dtype: int64
Dask Name: value-counts-agg, 4 tasks
%%timeit
dfsingle['Gender'].value_counts().compute()
9.2 s ± 98.3 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Reading and parsing the CSV is part of the task graph. It happens each time you compute.
dfsingle['Gender'].value_counts().visualize()
df['Gender'].value_counts().visualize()
Running a second time isn't any faster.
%%timeit
df['Gender'].value_counts().compute()
7.01 s ± 278 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Memory Persistence¶
You can tell Dask that an object (DataFrame/Array/any other intermediary result in the task graph) should be kept in memory.
gender = df['Gender'].persist() #this takes a while
gender
Dask Series Structure:
npartitions=14
object
...
...
...
...
Name: Gender, dtype: object
Dask Name: getitem, 14 tasks
%%timeit
gender.value_counts().compute()
66.1 ms ± 206 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
gendersingle = dfsingle['Gender'].persist() #this takes a while
%%timeit
gendersingle.value_counts().compute()
61.1 ms ± 172 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Reminder: File Formats¶
CSV is a really horrible file format for distributed processing.
- Need to infer types
- Need to parse text
- Need to read entire file to extract a single column
- Newlines within text entries may break dask read_csv
Alternatives
- parquet: Column oriented, binary, typed, compressed, chunked
- HDF5: hierarchical (like a file system), groups and datasets, binary, types
Parquet¶
df.to_parquet('hospital.parquet')
(None,)
!du -sh hospital.parquet/
1.3G hospital.parquet/
ls hospital.parquet/
_common_metadata part.10.parquet part.2.parquet part.6.parquet _metadata part.11.parquet part.3.parquet part.7.parquet part.0.parquet part.12.parquet part.4.parquet part.8.parquet part.1.parquet part.13.parquet part.5.parquet part.9.parquet
Where did the parts come from?
pdf = dd.read_parquet('hospital.parquet')
pdf
| Health Service Area | Hospital County | Operating Certificate Number | Facility ID | Facility Name | Age Group | Zip Code - 3 digits | Gender | Race | Ethnicity | Length of Stay | Type of Admission | Patient Disposition | Discharge Year | CCS Diagnosis Code | CCS Diagnosis Description | CCS Procedure Code | CCS Procedure Description | APR DRG Code | APR DRG Description | APR MDC Code | APR MDC Description | APR Severity of Illness Code | APR Severity of Illness Description | APR Risk of Mortality | APR Medical Surgical Description | Payment Typology 1 | Payment Typology 2 | Payment Typology 3 | Attending Provider License Number | Operating Provider License Number | Other Provider License Number | Birth Weight | Abortion Edit Indicator | Emergency Department Indicator | Total Charges | Total Costs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| npartitions=14 | |||||||||||||||||||||||||||||||||||||
| object | object | float64 | float64 | object | object | object | object | object | object | object | object | object | int64 | int64 | object | int64 | object | int64 | object | int64 | object | int64 | object | object | object | object | object | object | float64 | float64 | float64 | int64 | object | object | object | object | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
%%timeit
pdf['Gender'].value_counts().compute()
174 ms ± 824 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
gender = pdf['Gender'].persist()
%%timeit
gender.value_counts().compute()
64.2 ms ± 215 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
HDF5¶
import h5py
mouse = h5py.File('mouse_tabula_muris_10x_log1p_cpm.h5ad','r')
X = mouse['X']
This is a sparse matrix representation
$$54967×18099 = 994,847,733 >> 105,321,967$$import scipy
X = mouse['X']
M = scipy.sparse.csr_matrix((X['data'],X['indices'],X['indptr']))
M
<54967x18099 sparse matrix of type '<class 'numpy.float32'>' with 105321967 stored elements in Compressed Sparse Row format>
Dask¶
Any object that looks like an array can be converted to a Dask Array. However, note that the initial creation of the sparse matrix was done in scipy and was not parallel - the array must fit in memory.
sparse = da.from_array(M)
sparse
|
Dask¶
Dask has native support for hd5 dense arrays.
flat = da.from_array(X['data'])
flat
|
Scheduling¶
The local scheduler can execute computations using either threads or _processes.
Which do you think will be faster?
- which has faster communication?
- which has better concurrency?
%%timeit
x = np.random.uniform(low=0, high=10, size=(10000, 10000))
y = x**2
683 ms ± 6.31 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Threads¶
import dask
dask.config.set(scheduler='threads')
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x**2
y
|
%%timeit
y.compute()
256 ms ± 4.04 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Processes¶
dask.config.set(scheduler='processes')
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x**2
y
|
%%timeit
y.compute()
5.62 s ± 340 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
def pysum(arr):
tot = 0
for val in arr.flatten():
tot += val
return np.array(tot).reshape(1,1)
%%timeit
x = np.random.uniform(low=0, high=10, size=(10000, 10000))
pysum(x**2)
7.11 s ± 235 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Threads¶
dask.config.set(scheduler='threads')
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x**2
%%timeit
y.map_blocks(pysum).compute()
6.22 s ± 58.5 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Processes¶
dask.config.set(scheduler='processes')
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x**2
%%timeit
y.map_blocks(pysum).compute()
3.14 s ± 27.8 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
import numba
import numpy as np
@numba.jit(nopython=True, nogil=True)
def numba_pysum(arr):
tot = 0
for val in arr.flatten():
tot += val
return np.array(tot,dtype=arr.dtype).reshape(1,1)
Threads¶
dask.config.set(scheduler='threads')
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x**2
%%timeit
y.map_blocks(numba_pysum).compute()
211 ms ± 9.58 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Processes¶
dask.config.set(scheduler='processes')
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x**2
%%timeit
y.map_blocks(numba_pysum).compute()
3.3 s ± 58.3 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
Takeaway¶
For numpy/pandas heavy code or nogil numba code, use threads due to their low communication overhead, otherwise the Dask distributed scheduler is recommended, even if run on a single machine (more efficient implementation than the local processes scheduler).
from distributed import Client
import dask.array as da
client = Client(processes=False)
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x**2
%%timeit
y.map_blocks(numba_pysum).compute()
267 ms ± 12.6 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
%%timeit
y.map_blocks(pysum).compute()
6.37 s ± 18.1 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
client.close()
client = Client(processes=True)
x = da.random.uniform(low=0, high=10, size=(10000, 10000), chunks=(1000, 1000))
y = x**2
%%timeit
y.map_blocks(numba_pysum).compute()
239 ms ± 51.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
%%timeit
y.map_blocks(pysum).compute()
1.46 s ± 256 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)