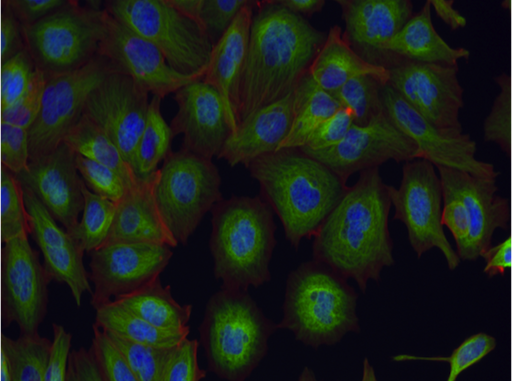
False color image of cells treated with an Aurora kinase inhibitor.
Assign6: Convolutional phenotyping
Checkpoint Friday, March 8, 11:59pm Friday, March 15, 11:59pm
Due Thursday, March 21, 11:59pm
SubmitFor this assignment you will use PyTorch to design and train a neural network to classify images of cells with respect to the mechanism of action of the molecule they were treated with.
Data
The source data comes from the Broad Institute. Cells from the MCF7 breast cancer cell line were treated with a variety of chemicals, labeled for DNA, F-actin, and B-tubulin, and imaged by fluorescent microscopy. The different compounds used to treat the cells have different mechanisms of action (how they effect the cell). The images are labeled based on the mechanism of action (MOA) of the compounds (labels provided by the Broad based on the scientific literature). Your task is to predict the mechanism of action from the fluorescent images. This would be useful, for example, in deducing the mechanism of action of new compounds. This is a multi-class classification problem.
| MOA | Train | Test |
|---|---|---|
| Actin_disruptors | 225 | 75 |
| Aurora_kinase_inhibitors | 540 | 180 |
| Cholesterol-lowering | 270 | 90 |
| DMSO | 4950 | 165 |
| DNA_damage | 405 | 135 |
| DNA_replication | 360 | 120 |
| Eg5_inhibitors | 540 | 180 |
| Epithelial | 330 | 110 |
| Kinase_inhibitors | 150 | 50 |
| Microtubule_destabilizers | 630 | 210 |
| Microtubule_stabilizers | 5355 | 178 |
| Protein_degradation | 315 | 105 |
| Protein_synthesis | 360 | 120 |
The counts for each MOA class are shown at the left. The training set is dominated by two classes, DMSO (the control - normal looking cells) and microtubule stabilizers. However, the test set is more balanced, reflecting the fact that we want a classifier to do well on all classes, not just the most dominant.
Training
To get you started, we are providing you with a fully functional CNN model:
train.py.
You should read through this code carefully, as you will be making significant changes to it.
This model does not necessarily have reasonable choices for the network architecture/hyperparameters.
Training
You can run the model locally on your own machine.$ ./train.py --train_data_dir /local/path/to/train Epoch 0 Step 0: loss = 332.424011 Epoch 0 Step 1: loss = 4112.622070 Epoch 0 Step 2: loss = 11168.931641 Epoch 0 Step 3: loss = 2756.125000 Epoch 0 Step 4: loss = 992.988586 Epoch 0 Step 5: loss = 712.976746 Epoch 0 Step 6: loss = 406.597198
By default, the script will fetch the training examples from Google Cloud. If you are planning on training locally (not on Google Cloud), you should download the entire training set (22GB tarball). Training on locally stored images will be substantially faster than fetching images from the network each time. You can specify the location of the training set on the commandline, as shown above.
When training on a cluster you will be able to accelerate training by first copying the training images to the local hard drive. In addition to the CSB cluster, you should have received an allocation (mscbio2066_2024s) on the University CRC cluster. Note that if you already have a CRC allocation you will need to set --account=mscbio2066_2024s to avoid using up your existing service units. A CRC quick reference guide is available here and more detailed documentation is here. The training data is available on the CRC cluster at /ihome/dkoes/dkoes/zfs/train and on the CSB cluster at /net/galaxy/home/koes/dkoes/data/train.tar. Make sure you copy the training data to the local scratch drive instead of reading the files from a network drive during training.
Cluster Etiquette Please read this document paying attention to the "Cluster Etiquette" and "Consideration of Others" sections.
In addition to the clusters, you can use your Google Cloud credits to spin up GPU enabled virtual machines. You could create a startup script that installs any necessary software/data and automatically starts a wandb agent to make running training runs as easy as launching the VM. When setting up a VM with limited storage space, rsync may be a better choice than downloading the tarball (since that requires twice the space):
mkdir train gsutil rsync -av gs://mscbio2066-data/trainimgs train/
A great way to easily make use of all the resources available is to use wandb sweeps to coordinate the launching of your training jobs. The clients, whether they be on CSB, CRC, or Google Cloud, will simply launch the wandb agent which will take care of launching your training script with different arguments. On a cluster you should use the --count 1 option since otherwise the agent will keep running even if there are no jobs to process. For example, on CSB your slurm script might be:
#!/bin/bash #SBATCH --job mscbio2066 #SBATCH --partition=dept_gpu #SBATCH --nodes=1 #SBATCH --gres=gpu:1 echo Running on `hostname` echo workdir $SLURM_SUBMIT_DIR module load cuda ~/.local/bin/wandb login #your identifier goes here# ~/.local/bin/wandb agent --count 1 #your sweep id goes here#
Optimization
The provided model, after training one epoch, has an accuracy of 11.5% on the test set. Your task is to improve these results.- Set up some sort of cross-validation. You should not rely on the test set to optimize your model (we don't provide it for you, you don't want to bog down the evaluation server, and it is bad practice to optimize hyperparameters on the final test set).
- Consider various forms of data augmentation to expand the training set. At the very least, consider modifying how batches are sampled to avoid overwhelming the network with dominant classes (balance your data). You may not expand the training set with entirely new images (e.g., additional images downloaded from the Broad), but you may generate variations of provided images (e.g., by rotating or flipping).
- Optimize the design of your network and training hyperparameters. Weights & Biases sweeps should be very useful for this. For your initial parameter search, use a reduced training and evaluation set and train for a limited number of iterations. Many hyperparameter sets will learn poorly or not at all and you do not want to spend a lot of time on them. Feel free to adapt previously published network architectures.
- Once you have a good set of hyperparameters and network architecture, train extensively on the full training set to create your final model.
Evaluation
Once you have a trained model you are happy with, you should submit a link to the model directory to the evaluation server (below). The link must be a world readable Google Storage location (gs://). You can copy your model output to a storage bucket with gsutil:gsutil acl ch -r -u AllUsers:R gs://bucket-name/ # set permissions gutil cp -r model.pth gs://bucket-name/ # copy dataIf gsutil isn't already installed, you can install it following these instructions. The model will be evaluated on the test set using the procedure in eval.py. Your model will be evaluated with PyTorch 2.2.0 and an NVIDIA TITAN RTX GPU (we assume the submitted model is trained on a GPU). We consider the overall accuracy and two methods of computing multi-class F1 (see here for more details). For reference, the best solution from previous years achieved a 97% accuracy.
Grading
Your grade will be $$grade = min(100, accuracy * 110)$$ For example, accuracies of 0.70, 0.80, and 0.90 will receive grades of 77, 88, and 99 respectively.Checkpoint
You must submit a working model with an accuracy greater than 0.15 by the checkpoint date or 10 points will be deducted from your final grade.Bonus
There is a 5 point bonus for achieving better than 97% accuracy.